Syntax Highlighting for Computational Biology.
Our goal is to make data intuitive for scientists and help you navigate and comprehend its significance. Currently supporting .sam, .vcf, .fasta, .fastq, .gtf, .bed, .pdb, .cwl, & more formats.
Tell us how bioSyntax can develop to help your workflow. (Survey ~5m)
INSTALL bioSyntax
Usage
bioSyntax integrates seamlessly with vim, less, gedit, & sublime to automatically recognize your favorite biological file formats. To gain the most insight from your data, read our brief bioSyntax Manual.
Large data can also be directly piped into less with sam-less, vcf-less, …, xyz-less commands.
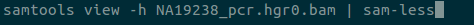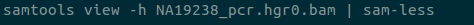
Collaborate
We’re actively developing bioSyntax; we’d love to hear your comments, feedback and suggestions for further development. Drop us a line on github or email.
If you’d like to help out, have intimate understanding of a scientific data-type, or are looking for a fun design / optimization problem check the development page.
Updates
2018-08-22 - bioSyntax publication
- Our manuscript has been accepted for publication in BMC Bioinformatics
2018-08-13 - bioSyntax v1.0 Release
- Syntax highlighting for FASTA, FASTQ, CWL, BED, GTF, PDB, PML, SAM, VCF file formats
- Available for:
less,vim,gedit,sublimeandVScode - Updated installer script + packaged documentation
- Welcome Dylan Aissi + Li Yao on as developers
2018-04-11 - bioSyntax v0.1-beta4 + Development Survey
- We’re working on a user experience survey to focus development.
- New Syntax
vim-nexus - New Syntax
vim-pml&sublime-pml(Pymol Script Language) - New Syntax
cwl-sublimeandcwl-vim(Common Workflow Language) - Add
bam-lessalias forsam-lessby default - Removed
sudorequirements for linux-gedit & linux-less installations - Alternative Syntax:
vim_fasta-ORF: Syntax for finding and highlighting Open Reading Frames
Thanks to
- Luis Carvalho for
vim-nexus - @manabuishii for
cwl-sublimeandcwl-vim - @speleo3 for
vim-pml& @bbarad forsublime-pml